Gene genealogies
The standard model for genetic evolution is the coalescent process. It is a stochastic model for how the ancestral lines of individuals sampled from a biological population form its gene genealogy. This genealogy in turn determines the degree of genetic variation observed in the sample. Larger variation allows the population to adapt more easily to a changing environment. In the limit of large population sizes, coalescent processes and related models can be analysed using diffusion approximations, not unlike models for the paths of particles in turbulent aerosols.
Using mathematical analysis of coalescent processes and computer simulations of related models we analysed how gene genealogies and genetic variation are affected by demographic factors (population-size changes, and spatial population structure and migration), see Figure 1.
This led to collaborations with colleagues from the Tjärnö Marine Laboratory at the University of Gothenburg, where we contributed to the analysis of the population genetics of the marine snail Littorina saxatilis.
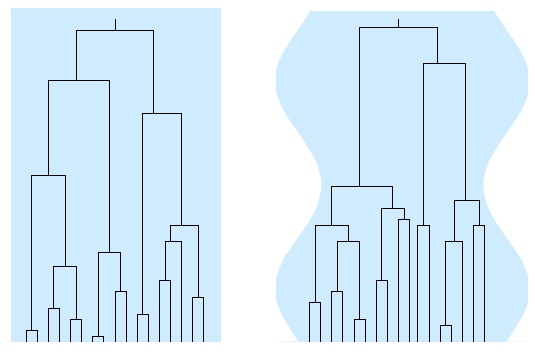
Figure 1. Gene genealogies in a population of constant size (left), and in a population with a time-varying population size. From Ref. [10].
[1] Clines on the seashore: The genomic architecture underlying rapid divergence in the face of gene flow
AM Westram, M Rafajlović, P Chaube, R Faria, T Larsson, M Panova, Mark Ravinet, Anders Blomberg, Bernhard Mehlig, Kerstin Johannesson & Roger Butlin, Evolution Letters 2 (2018), 297-309
[2] Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow
M Ravinet, R Faria, RK Butlin, J Galindo, N Bierne, M Rafajlović, MAF Noor, B Mehlig & AM Westram, Journal of Evolutionary Biology 30 (2017) 1450-1477
[3] Neutral processes forming large clones during colonization of new areas
M Rafajlović, D Kleinhans, C Gulliksson, J Fries, D Johansson, A Ardehed, L Sundqvist, RT Pereyra, B Mehlig, PR Jonsson & K Johannesson, Journal of Evolutionary Biology 30 (2017), 1544-1560
[4] A universal mechanism generating clusters of differentiated loci during divergence‐with‐migration
M Rafajlović, A Emanuelsson, K Johannesson, RK Butlin & B Mehlig, Evolution 70 (2016) 1609-1621
[5] Demography-adjusted tests of neutrality based on genome-wide SNP data
M Rafajlović, A Klassmann, A Eriksson, T Wiehe & B Mehlig, Theoretical Population Biology 95 (2014) 1-12
[6] The emergence of the rescue effect from explicit within-and between-patch dynamics in a metapopulation
A Eriksson, F Elías-Wolff, B Mehlig &A Manica, Proceedings of the Royal Society B: Biological Sciences 281 (2014) 20133127
[7] The effect of multiple paternity on genetic diversity of small populations during and after colonisation
M Rafajlović, A Eriksson, A Rimark, S Hintz-Saltin, G Charrier, M Panova, Carl André, Kerstin Johannesson & Bernhard Mehlig, PloS ONE 8 (2013) e75587
[8] Evolutionary branching in a stochastic population model with discrete mutational steps
S Sagitov, B Mehlig, P Jagers & V Vatutin, Theoretical Population Biology 83 (2013) 145-154
[9] Linkage disequilibrium under recurrent bottlenecks
E Schaper, A Eriksson, M Rafajlovic, S Sagitov & B Mehlig, Genetics 190 (2012) 217-229
[10] The total branch length of sample genealogies in populations of variable size
A Eriksson, B Mehlig, M Rafajlovic & S Sagitov, Genetics 186 (2010) 601-611
[11] Extreme female promiscuity in a non-social invertebrate species
M Panova, J Boström, T Hofving, T Areskoug, A Eriksson, B Mehlig, Tuuli Mäkinen, Carl Andre & Kerstin Johannesson, PLoS ONE 5 (2010) e9640
[12] Multiple paternity: determining the minimum number of sires of a large brood
A Eriksson, B Mehlig, M Panova, C André & K Johannesson, Molecular Ecology Resources 10 (2010), 282-291
[13] Sequential Markov coalescent algorithms for population models with demographic structure
A Eriksson, B Mahjani & B Mehlig, Theoretical Population Biology 76 (2009) 84-91
[14] An accurate model for genetic hitchhiking
A Eriksson, P Fernstrom, B Mehlig & S Sagitov, Genetics 178 (2008) 439-451
[15] On the effect of fluctuating recombination rates on the decorrelation of gene histories in the human genome
A Eriksson & B Mehlig, Genetics 169 (2005) 1175-1178