DNA in nanochannels
In genome mapping experiments, long DNA molecules are stretched by confining them to nanochannels. When the DNA is fully extended, the distances X along the channel axis between sequence-specific fluorescent labels provide genetic fingerprints. However, it is a challenge to make the channels narrow enough so that the DNA molecules are fully stretched. In practice their conformations may form hairpins that change the label spacings along the channel axis (Figure).
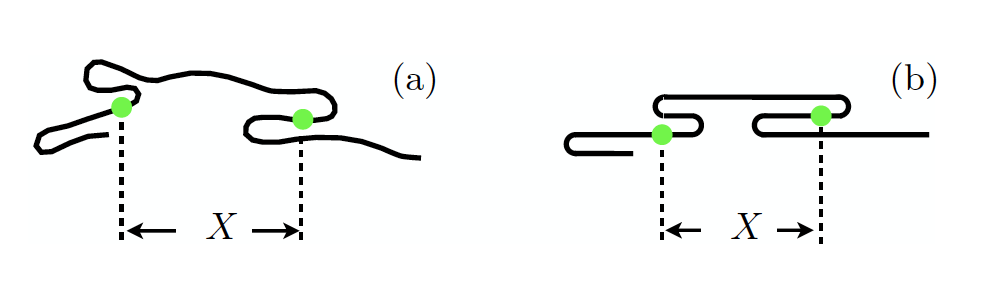
Figure. (a) Conformation of a confined DNA molecule. The distance along the channel axis between the two fluorescent labels (green) is denoted by X. The conformation shown exhibits two S-hairpins and a C-hairpin at the left end. (b) Representation within the telegraph model. From Ref. [3].
How frequent are such hairpins? Not only had this question remained unanswered until recently, but even the most basic scaling predictions for the conformations of confined DNA are at odds with experiments measuring DNA extension in nanochannels.
We developed a theory based on a stochastic, weakly self-avoiding, one-dimensional telegraph process that collapses experimental data and simulation results onto a single master curve throughout the experimentally relevant region of parameter space [4], and that explains the mechanisms at play [2,3,12].
Based on our understanding of confined DNA conformations we calculated how confinement affects the melting of the DNA, the separation of its strands [8,9], together with colleagues at Lund University.
During the past years, we contributed to the analysis of experimental results on the conformation and dynamics of nanoconfined DNA [1,5,7,11].
[1] Stochastic unfolding of nanoconfined DNA: Experiments, model and Bayesian analysis
J Krog, M Alizadehheidari, E Werner, SK Bikkarolla, JO Tegenfeldt, Bernhard Mehlig, Michael A Lomholt, Fredrik Westerlund & Tobias Ambjörnsson, J. Chem. Phys. 149 (2018) 215101
[2] Hairpins in the conformations of a confined polymer
E Werner, A Jain, A Muralidhar, K Frykholm, T St Clere Smithe, J Fritzsche, F Westerlund, KD Dorfman & B Mehlig, Biomicrofluidics 12 (2018) 024105
[3] Distribution of label spacings for genome mapping in nanochannels
D. Ödman, E. Werner, K. D. Dorfman, C. R. Doering & B. Mehlig, Biomicrofluidics 12 (2018) 034115
[4] One-Parameter Scaling Theory for DNA Extension in a Nanochannel
E Werner, GK Cheong, D Gupta, KD Dorfman & B Mehlig, Phys. Rev. Lett. 119 (2017) 268102
[5] Visualizing the nonhomogeneous structure of RAD51 filaments using nanofluidic channels
LH Fornander, K Frykholm, J Fritzsche, J Araya, P Nevin, E Werner, Ali Çakır, Fredrik Persson, Edwige B Garcin, Penny J Beuning, Bernhard Mehlig, Mauro Modesti & Fredrik Westerlund, Langmuir 32 (2016) 8403-8412
[6] Finite-size corrections for confined polymers in the extended de Gennes regime
TSC Smithe, V Iarko, A Muralidhar, E Werner, KD Dorfman, B Mehlig, Physical Review E 92 (2015) 062601
[7] Extension of nanoconfined DNA: Quantitative comparison between experiment and theory
V Iarko, E Werner, LK Nyberg, V Müller, J Fritzsche, T Ambjörnsson, JP Beech, JO Tegenfeldt, K Mehlig, F Westerlund & B Mehlig, Physical Review E 92 (2015) 062701
[8] How nanochannel confinement affects the DNA melting transition within the Poland-Scheraga model
M Reiter-Schad, E Werner, JO Tegenfeldt, B Mehlig & T Ambjörnsson, J. Chem. Phys. 143 (2015) 09B616_1
[9] Model for melting of confined DNA
E Werner, M Reiter-Schad, T Ambjörnsson & B Mehlig, Physical Review E 91 (2015) 060702
[10] Scaling regimes of a semiflexible polymer in a rectangular channel
E Werner & B Mehlig, Physical Review E 91 (2015) 050601
[11] Nanoconfined circular and linear DNA: Equilibrium conformations and unfolding kinetics
M Alizadehheidari, E Werner, C Noble, M Reiter-Schad, LK Nyberg, Joachim Fritzsche, Bernhard Mehlig, Jonas O Tegenfeldt, Tobias Ambjörnsson, Fredrik Persson & Fredrik Westerlund, Macromolecules 48 (2015) 871-878
[12] Confined polymers in the extended de Gennes regime
E Werner & B Mehlig, Physical Review E 90 (2014) 062602
[13] Monomer distributions and intrachain collisions of a polymer confined to a channel
E Werner, F Westerlund, JO Tegenfeldt & B Mehlig, Macromolecules 46 (2013) 6644-6650
[14] Orientational correlations in confined DNA
E Werner, F Persson, F Westerlund, JO Tegenfeldt & B Mehlig, Physical Review E 86 (2012) 041802